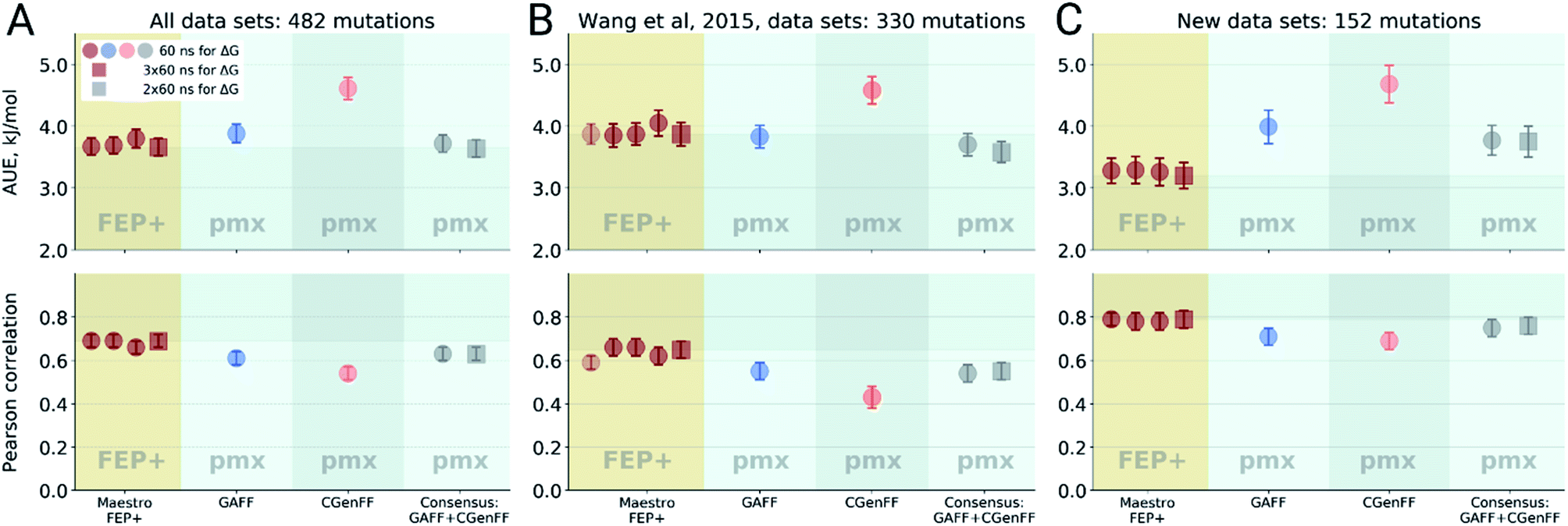
Precise force-field-based calculations of octanol-water partition coefficients for the SAMPL7 molecules | SpringerLink
molecular dynamics - Alternative to CGenFF for generating large ligand topology - Matter Modeling Stack Exchange

Molecules | Free Full-Text | Comparing Dimerization Free Energies and Binding Modes of Small Aromatic Molecules with Different Force Fields | HTML
GitHub - jandom/paramchem: Python scrapper for paramche/cgenff server, small molecule parametrization

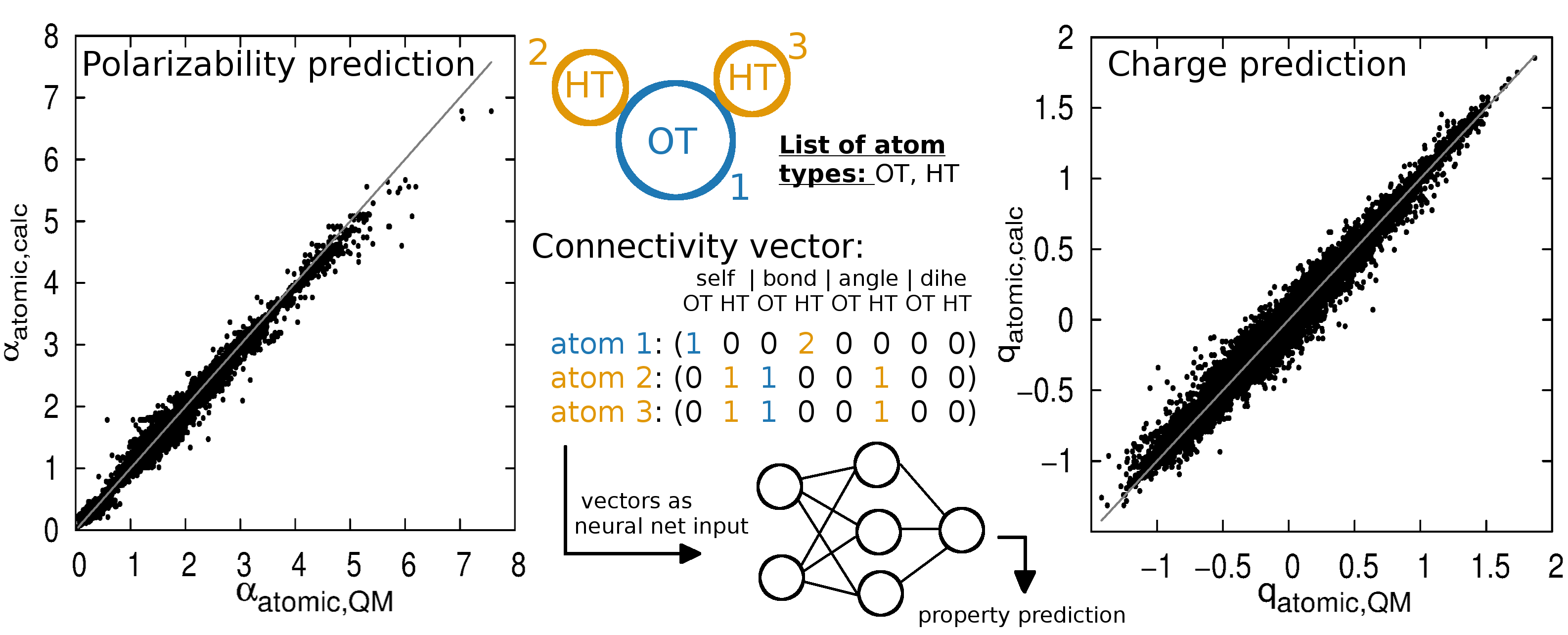
Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing | Semantic Scholar
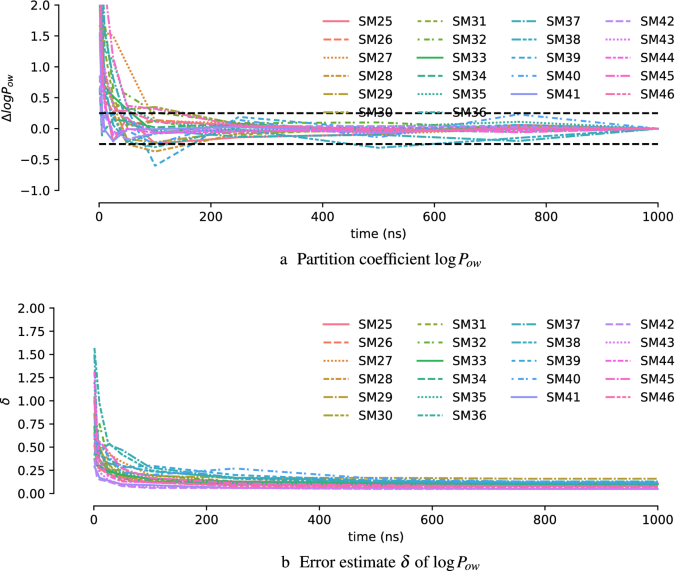
Precise force-field-based calculations of octanol-water partition coefficients for the SAMPL7 molecules | SpringerLink
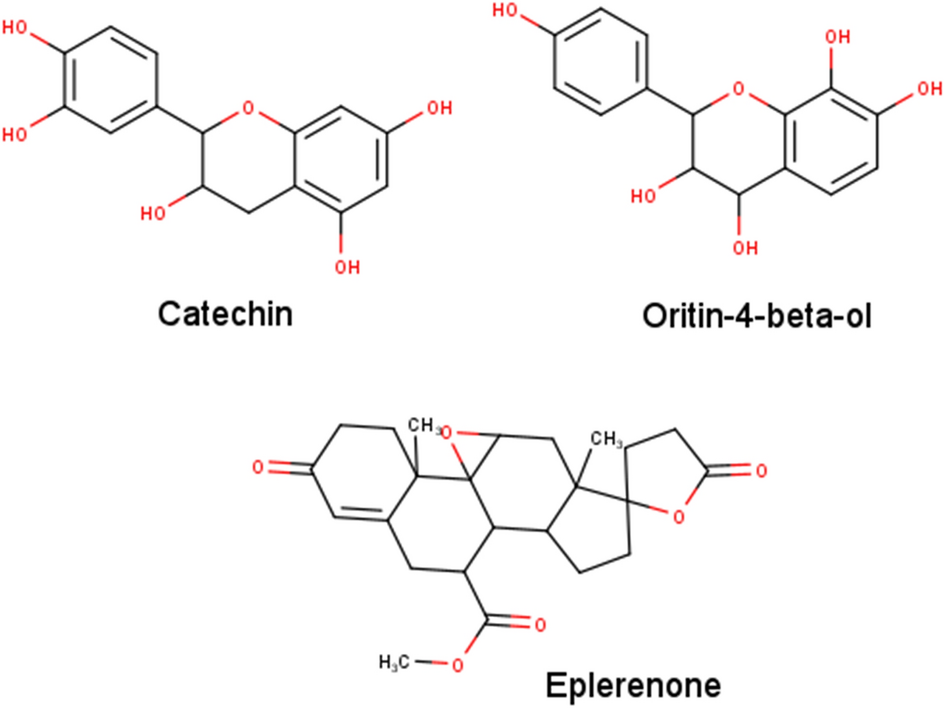
A scaffolded approach to unearth potential antibacterial components from epicarp of Malaysian Nephelium lappaceum L. | Scientific Reports

Additive CHARMM force field for naturally occurring modified ribonucleotides - Xu - 2016 - Journal of Computational Chemistry - Wiley Online Library
![Simultaneous molecular docking of different ligands to His6-tagged organophosphorus hydrolase as an effective tool for assessing their effect on the enzyme [PeerJ] Simultaneous molecular docking of different ligands to His6-tagged organophosphorus hydrolase as an effective tool for assessing their effect on the enzyme [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2019/7684/1/fig-3-full.png)
Simultaneous molecular docking of different ligands to His6-tagged organophosphorus hydrolase as an effective tool for assessing their effect on the enzyme [PeerJ]
![In silico characterization and homology modeling of cytosolic APX gene predicts novel glycine residue modulating waterlogging stress response in pigeon pea [PeerJ] In silico characterization and homology modeling of cytosolic APX gene predicts novel glycine residue modulating waterlogging stress response in pigeon pea [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/10888/1/fig-2-full.png)
In silico characterization and homology modeling of cytosolic APX gene predicts novel glycine residue modulating waterlogging stress response in pigeon pea [PeerJ]

CHARMM force field parameters for 2′-hydroxybiphenyl-2-sulfinate, 2-hydroxybiphenyl, and related analogs - ScienceDirect